31+ Extinction Coefficient Calculator
Web This video explains how to work out the extinction coefficient for a solution from a standard curve. E MGdn-HCl aE MTyr bE MTrp cE MCys Where abc are the number of tyrosine trytophan and cystine residues per mole of protein and E.

Pdf Amyloid Proteins Miguel Calero Academia Edu
1 OD 260 Unit 50 μgml for dsDNA 1 OD 260 Unit 40 μgml ssRNA 1 OD 260 Unit 33 μgml.

. Web Protein Extinction Coefficients and Concentration Calculation. Web Our standard extinction coefficient calculation procedure consists of. Web Protein extinction coefficient calculation in high-quality scientific databases and software tools using Expasy the Swiss Bioinformatics Resource Portal.
Web DNA Concentration Calculator. Web extinction coefficient that will be used to calculate the protein concentration based on the sample absorbance at 205 nm. Web The Molar extinction coefficient formula is defined as a measure of how strongly a chemical species or substance absorbs light at a particular wavelength and is.
Web ProtParam References Documentation is a tool which allows the computation of various physical and chemical parameters for a given protein stored in Swiss-Prot or TrEMBL or. Web Formula concentration ugml OD 260 x conversion factor conversion factors. The concentration of DNA in solution can be determined by substituting the molecular weight extinction coefficient and λ max into a derived form of.
Web The calculations is as follows. Web Sample Type Option Extinction Coefficient BSA 667 IgG 137 Lysozyme 246 1 Abs 1 mgmL Default general reference setting E1 User-entered mass extinction coefficient. Web The extinction coefficient is the absorbance divided by the concentration and the pathlength according to Beers Law epsilon.
This quantity is a bit tricky despite being a rather simple addition. First we determine the exact quantity of proteinantibody by our amino acid analysis service. Show more Generating Standard Curve and Determination of Protein.
Web The extinction coefficient of a material describes how strongly it absorbs light. Web percent solution extinction coefficients ie one must convert from 10 mgml units to 1 mgml concentration units. Von Hippel presented a method for calculation accurate to 5 in most cases molar.
Percent 10 concentration in mgml. ε 205 31 method Scopes method2 Other custom. Web AAT Bioquest Protein Concentration Calculator The concentration of Protein in solution can be determined by substituting the molecular weight extinction coefficient and λ max.
Web Oligo Extinction Coefficient Calculator This form can be used in two different ways. Gill and Peter H. You can either enter your sequence lowerupper case spaces OK or enter the number of.

Extinction Coefficient Calculator Calculator Academy
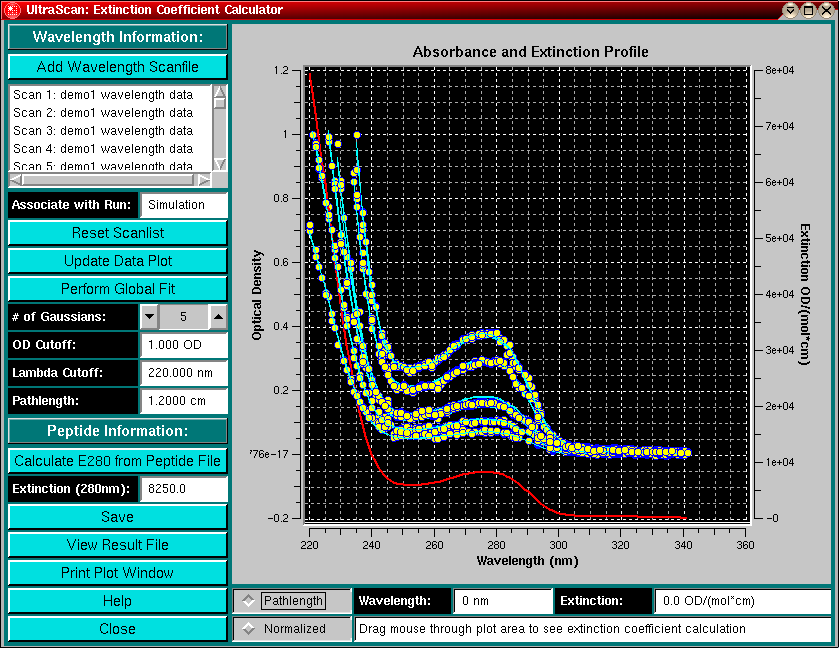
Protein Extinction Profile Calculation
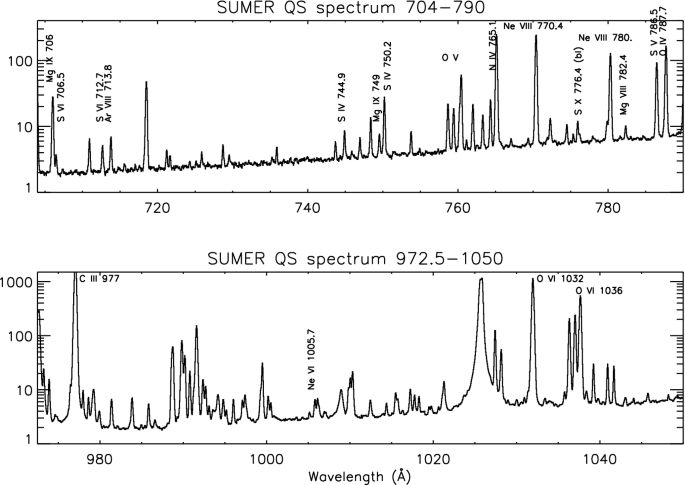
Solar Uv And X Ray Spectral Diagnostics Springerlink

Visible Light Mediated Modification And Manipulation Of Biomacromolecules Chemical Reviews

Vegetation Indices X5675602的博客 Csdn博客

Atmospheric Propagation In The Uv Visible Ir And Mm Wave Region And Related Systems Aspects Pdf Aerosol Wound

Elimination Of Interference From Water In Kbr Disk Ft Ir Spectra Of Solid Biomaterials By Chemometrics Solved With Kinetic Modeling Sciencedirect
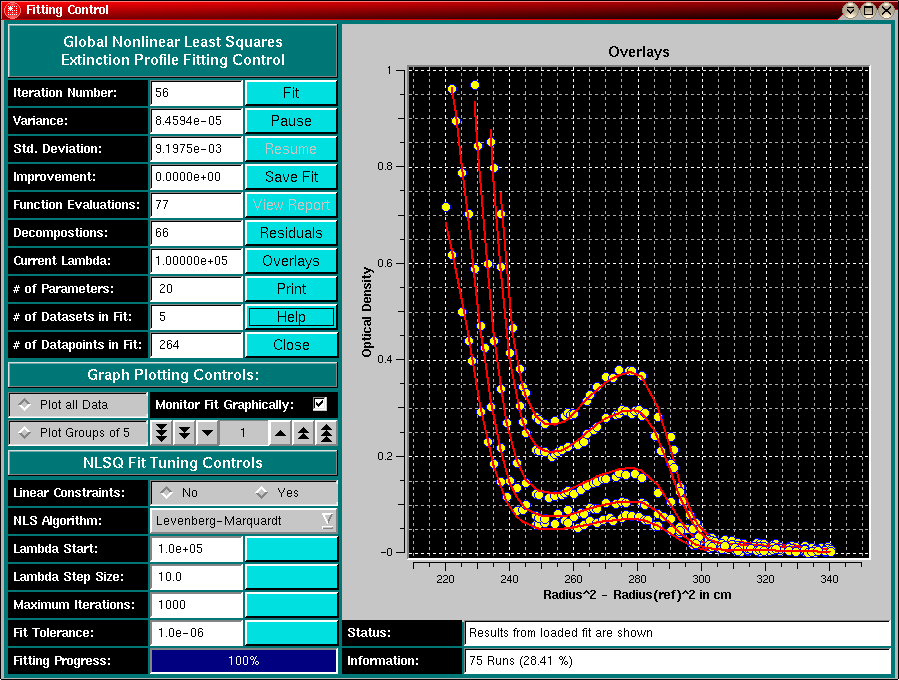
Protein Extinction Profile Calculation

Absorption Emission With Molar Extinction Coefficient At 1 10 6 M Download Table

Roles Of Bromine Radicals And Hydroxyl Radicals In The Degradation Of Micropollutants By The Uv Bromine Process Environmental Science Technology

Extinction Coefficient Calculation Accurate Quantification

User Moira M Esson Notebook Chem 571 2013 09 04 Openwetware
Nick Anthis Protein Parameter Calculator
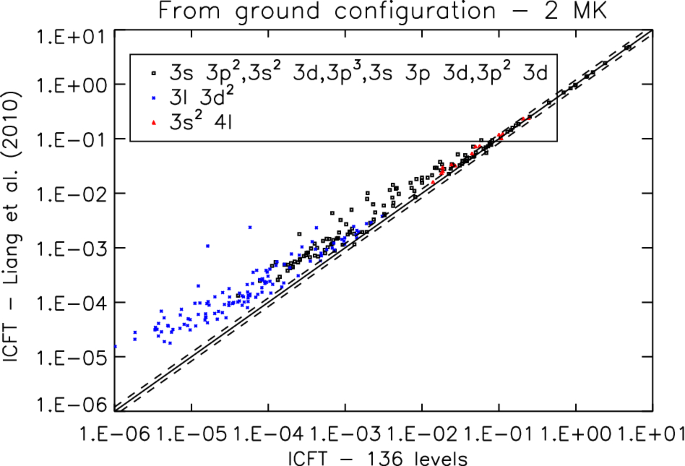
Solar Uv And X Ray Spectral Diagnostics Springerlink
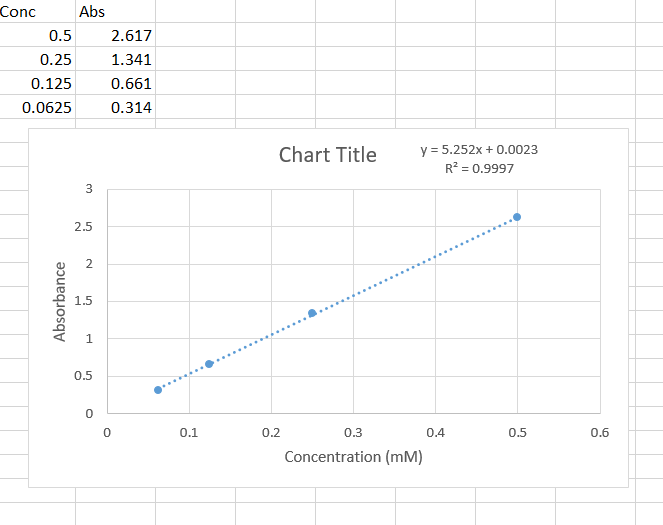
Solved Calculate The Extinction Coefficient Using The Chegg Com
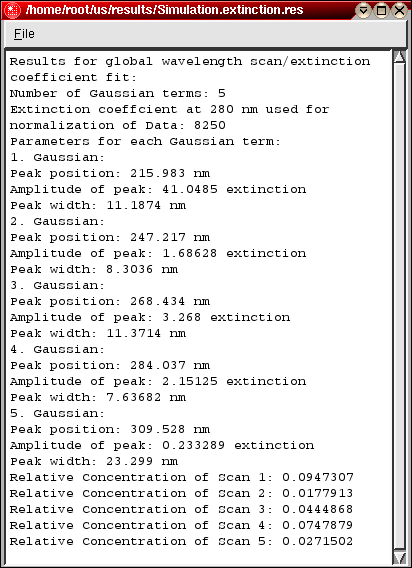
Protein Extinction Profile Calculation

Extinction Coefficient Calculation Accurate Quantification